Spatial-CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level
Deng et al.,
Science, 2022
Publications
Manuscripts under review
Alev Baysoy, Xiaolong Tian, Feifei Zhang, Paul Renauer, Zhiliang Bai, Hao Shi, Haikuo Li, Bo Tao, Mingyu Yang, Archibald Enninful, Fu Gao, Guangchuan Wang, Wanqiu Zhang, Thao Tran, Nathan Heath Patterson, Shuozhen Bao, Chuanpeng Dong, Shan Xin, Mei Zhong, Sherri Rankin, Cliff Guy, Yan Wang, Jon P. Connelly, Shondra M. Pruett- Miller, Hongbo Chi, Sidi Chen*, Rong Fan*, Spatially Resolved Panoramic in vivo CRISPR Screen via Perturb-DBiT, under review (2024)
Spatially resolved m6A profiling using m6A-ARTR-DBiT, Yu Xiao, Zhiliang Bai, Chang Ye, Bo Tao, Zhong Zheng, Yan-Ming Chen, Zhongyu Zou, Zhuoning Zou, Yuhang Fan, Yun Gao, Rong Fan*, Chuan He*, under revision (2025)
Negin Farzad, Archibald Enninful, Yao Lu, Anthony Fung, Yajuan Li, Fabio Parisi, Mingyu Yang, Francesco Strino, Junchen Yang, Mei Zhong, Joseph Cunningham, Zhiliang Bai, Haikuo Li, Fang Wang, Michael Stankewich, Dongjoo Kim, Mingze Dong, Keyi Li, Yu Mi Kwon, Liang Chen, Ljiljana Pasa-Tolic, Joseph Craft, Lingyan Shi, Yuval Kluger, Stephanie Halene, Mina L. Xu, Rong Fan, Human Lymph Node Cellular Senescence Atlas Reveals Age-Dependent Alteration in Germinal Center B Cell Function and Niches, under revision (2024)
Yajuan Li, Zhaojun Zhang, Archibald Enninful, Jungmin Nam, Xiaoyu Qin, Jorge Villazon, Negin Fazard, Anthony A. Fung, Mina L. Xu, Hongje Jang, Nancy R. Zhang, Rong Fan*, Zongming Ma*, Lingyan Shi*, All-Optical Multimodal Mapping of Single Cell Type-Specific Metabolic Activities via REDCAT, under revision (2024)
Fu Gao, Yao Lu, Irbaz Hameed, Sufang Li, Mei Zhong, Archibald Enninful, Xing Lou, Mingze Dong, Bo Tao, Di Zhang, Zhiliang Bai, Dongjoo Kim, Shuozhen Bao, Mingyu Yang, Fang Wang, Markus Krane, George Tellides, Yang Liu*, Arnar Geirsson*, and Rong Fan*, A single-cell and spatial atlas of human and murine mitral valve and valvular diseases, under revision (2024)
Adrian C. Johnston, Gretchen M. Alicea, Cameron C. Lee, Payal V. Patel, Eban A. Hanna, Eduarda Vaz, André Forjaz, Zeqi Wan, Praful R. Nair, Yeongseo Lim, Tina Chen, Wenxuan Du, Dongjoo Kim, Tushar D. Nichakawade, Vito W. Rebecca, Challice L. Bonifant, Rong Fan, Ashley L. Kiemen, Pei-Hsun Wu, Denis Wirtz*, Engineering self-propelled tumor-infiltrating CAR T cells using synthetic velocity receptors, under revision (2024).
Selected Publications
Archibald Enninful, Zhaojun Zhang, Dmytro Klymyshyn, Matthew Ingalls, Mingyu Yang, Hailing Zong, Zhiliang Bai, Negin Farzad, Graham Su, Alev Baysoy, Jungmin Nam, Yao Lu, Shuozhen Bao, Siyan Deng, Nancy R. Zhang, Oliver Braubach, Mina L. Xu*, Zongming Ma*, Rong Fan*, Spatially Resolved Multi-Omics Profiling On Same Tissue Sections, Nature Methods (2025)
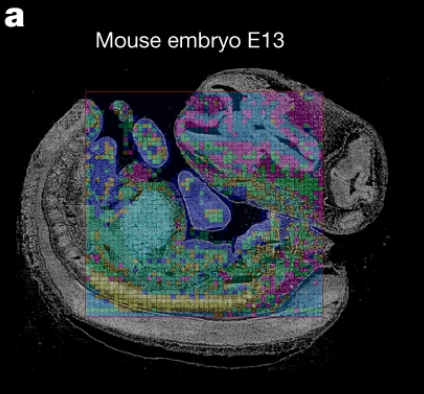
Di Zhang, Leslie A Rubio Rodríguez-Kirby, Yingxin Lin, Mengyi Song, Li Wang, Lijun Wang, Shigeaki Kanatani, Tony Jimenez-Beristain, Yonglong Dang, Mei Zhong, Petra Kukanja, Shaohui Wang, Xinyi Lisa Chen, Fu Gao, Dejiang Wang, Hang Xu, Xing Lou, Yang Liu, Jinmiao Chen, Nenad Sestan, Per Uhlén, Arnold Kriegstein, Hongyu Zhao, Gonçalo Castelo-Branco*, and Rong Fan*, Spatial dynamics of mammalian brain development and neuroinflammation by multimodal tri-omics mapping, Nature (2025)
Haikuo Li, Shuozhen Bao, Negin Farzad, Xiaoyu Qin, Anthony A. Fung, Di Zhang, Zhiliang Bai, Bo Tao, Rong Fan*, Spatially resolved genome-wide joint profiling of epigenome and transcriptome (spatial-ATAC-RNA-seq and spatial- CUT&Tag-RNA-seq), Nature Protocols, in press (2025)
Rong Fan*, Integrative Spatial Protein Profiling with Multi-Omics, Nature Methods, 21, 2223-2225 (2024).
Zhiliang Bai, Dingyao Zhang, Yan Gao, Bo Tao, Shuozhen Bao, Archibald Enninful, Daiwei Zhang, Graham Su, Xiaolong Tian, Ningning Zhang, Yang Liu, Mark Gerstein, Mingyao Li*, Yi Xing*, Jun Lu*, Mina Xu*, & Rong Fan*, Spatially Exploring RNA Biology in Archival Formalin-Fixed Paraffin-Embedded Tissues via Patho-DBiT, Cell 187, 6760–6779 (2024). (Cover Highlight)
Zhiliang Bai, Bing Feng, Susan E. McClory, Beatriz Coutinho de Oliveira, Caroline Diorio, Céline Gregoire, Bo Tao, Luojia Yang, Ziran Zhao, Lei Peng, Giacomo Sferruzza, Liqun Zhou, Xiaolei Zhou, Jessica Kerr, Alev Baysoy, Graham Su, Mingyu Yang, Pablo G. Camara, Sidi Chen, Li Tang, Carl H. June, J. Joseph Melenhorst, Stephan A. Grupp, and Rong Fan*. Single-cell CAR T atlas reveals type-2 function in 8-year leukemia remission, Nature, 634, 702–711 (2024).
Bing Feng , Zhiliang Bai, Xiaolei Zhou, Yang Zhao, Yu-Qing Xie, Xinyi Huang, Yang Liu, Tom Enbar, Rongrong Li, Yi Wang, Min Gao, Lucia Bonati, Mei-Wen Peng, Weilin Li, Bo Tao, Mélanie Charmoy, Werner Held, J. Joseph Melenhorst, Rong Fan*, Yugang Guo*, and Li Tang*, A type 2 cytokine Fc–IL-4 revitalizes exhausted CD8 + T cells against cancer, Nature, 634, 712–720 (2024).
Negin Farzad, Archie Enninful, Shuozhen Bao, Di Zhang, Yanxiang Deng, and Rong Fan*. Spatially resolved epigenome sequencing via Tn5 transposition and deterministic DNA barcoding in tissue. Nature Protocols 19, 3389– 3425 (2024).
Yahui Long, Kok Siong Ang, Raman Sethi, Sha Liao, Yang Heng, Lynn van Olst, Shuchen Ye, Chengwei Zhong, Hang Xu, Di Zhang, Immanuel Kwok, Nazihah Husna, Min Jian, Lai Guan Ng, Ao Chen, Nicholas RJ Gascoigne, David Gate, Rong Fan, Xun Xu, Jinmiao Chen, Deciphering spatial domains from spatial multi-omics with SpatialGlue, Nature Methods, (2024). doi: 10.1038/s41592-024-02316-4
Vidyani Suryadevara, … Rong Fan, … SenNet consortium, SenNet recommendations for detecting senescent cells in different tissues, Nature Reviews Molecular Cell Biology, (2024) doi: 10.1038/s41580-024-00738-8
Praful R Nair, Ludmila Danilova, Estibaliz Gómez-de-Mariscal, Dongjoo Kim, Rong Fan, Arrate Muñoz-Barrutia, Elana J Fertig, Denis Wirtz, MLL1 regulates cytokine-driven cell migration and metastasis, Science Advances 10, eadk0785 (2024).
Di Zhang, Yanxiang Deng, Petra Kukanja, Eneritz Agirre, Marek Bartosovic, Mingze Dong, Cong Ma, Sai Ma, Graham Su, Shuozhen Bao, Yang Liu, Yang Xiao, Gorazd B. Rosoklija, Andrew J. Dwork, J. John Mann, Kam W. Leong, Maura Boldrini, Liya Wang, Maximilian Haeussler, Benjamin J. Raphael, Yuval Kluger, Gonçalo Castelo- Branco, Rong Fan*, Spatial epigenome-transcriptome co-profiling of mammalian tissues, Nature, 616, 113–122 (2023)
Yang Liu, Marcello DiStasio, Graham Su, Hiromitsu Asashima, Archibald Enninful, Xiaoyu Qin, Yanxiang Deng, Pino Bordignon, Marco Cassano, Mary Tomayko, Mina Xu, Stephanie Halene, Joseph E. Craft, David Hafler, and Rong Fan*, High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq, Nature Biotechnology, 41, 1405–1409 (2023).
Alev Baysoy, Zhiliang Bai, Rahul Satija, Rong Fan*, The technological landscape and applications of single-cell multi-omics, Nature Reviews Molecular Cell Biology, 24, 695–713 (2023) (cover highlight)
Yanxiang Deng, Zhiliang Bai, Rong Fan*, Microtechnologies for single-cell and spatial multi-omics, Nature Reviews Bioengineering, 1, 769–784 (2023)
Dongjoo Kim, Giulia Biancon, Zhiliang Bai, Jennifer VanOudenhove, Yuxin Liu, Shalin Kothari, Lohith Gowda, Jennifer M Kwan, Nicholas Carlos Buitrago‐Pocasangre, Nikhil Lele, Hiromitsu Asashima, Michael K Racke, JoDell E Wilson, Tara S Givens, Mary M Tomayko, Wade L Schulz, Erin E Longbrake, David A Hafler, Stephanie Halene, Rong Fan*, Small Methods, 7, 2300594 (2023).
Rong Fan* and Fay Lin*, Spatial Delivery, GEN Biotechnology, 2 (5), 331-332 (2023)
Hiromitsu Asashima, Dongjoo Kim, Kaicheng Wang, Nikhil Lele, Nicholas C Buitrago-Pocasangre, Rachel Lutz, Isabella Cruz, Khadir Raddassi, William E Ruff, Michael K Racke, JoDell E Wilson, Tara S Givens, Alba Grifoni, Daniela Weiskopf, Alessandro Sette, Steven H Kleinstein, Ruth R Montgomery, Albert C Shaw, Fangyong Li, Rong Fan, David A Hafler, Mary M Tomayko, Erin E Longbrake , Prior cycles of anti-CD20 antibodies affect antibody responses after repeated SARS-CoV-2 mRNA vaccination. JCI insight 8(16), e168102, (2023)
B Tao, H Yang, R Fan*, Seeing the Action of Lipid Droplets in Aging and Longevity, GEN Biotechnology, 2 (4), 287-289 (2023).
Rong Fan*, Mapping RNA translation, Science 380 (6652), 1321-1322 (2023).
Alexandra S Piotrowski-Daspit, Christina Barone, Chun-Yu Lin, Yanxiang Deng, Douglas Wu, Thomas C Binns, Emily Xu, Adele S Ricciardi, Rachael Putman, Alannah Garrison, Richard Nguyen, Anisha Gupta, Rong Fan, Peter M Glazer, W Mark Saltzman, Marie E Egan, In vivo correction of cystic fibrosis mediated by PNA nanoparticles, Science Advances, 8(40), eabo0522 (2022).
Andrew J Dunbar, Dongjoo Kim, Min Lu, Mirko Farina, Robert L Bowman, Julie L Yang, Young Park, Abdul Karzai, Wenbin Xiao, Zach Zaroogian, Kavi O’Connor, Shoron Mowla, Francesca Gobbo, Paola Verachi, Fabrizio Martelli, Giuseppe Sarli, Lijuan Xia, Nada Elmansy, Maria Kleppe, Zhuo Chen, Yang Xiao, Erin McGovern, Jenna Snyder, Aishwarya Krishnan, Corrine Hill, Keith Cordner, Anouar Zouak, Mohamed E Salama, Jayden Yohai, Eric Tucker, Jonathan Chen, Jing Zhou, Timothy McConnell, Anna R Migliaccio, Richard Koche, Raajit Rampal, Rong Fan*, Ross L Levine*, Ronald Hoffman*, CXCL8/CXCR2 signaling mediates bone marrow fibrosis and is a therapeutic target in myelofibrosis, Blood, 141 (20): 2508–2519 (2023).
Yoon-A Kang, Hyojung Paik, Si Yi Zhang, Jonathan J Chen, Oakley C Olson, Carl A Mitchell, Amelie Collins, James W Swann, Matthew R Warr, Rong Fan, Emmanuelle Passegué, Secretory MPP3 reinforce myeloid differentiation trajectory and amplify myeloid cell production, Journal of Experimental Medicine, 220 (8), e20230088 (2023).
Ann T Chen, Yang Xiao, Xiangjun Tang, Mehdi Baqri, Xingchun Gao, Melanie Reschke, Wendy C Sheu, Gretchen Long, Yu Zhou, Gang Deng, Shenqi Zhang, Yanxiang Deng, Zhiliang Bai, Dongjoo Kim, Anita Huttner, Russell Kunes, Murat Günel, Jennifer Moliterno, W Mark Saltzman, Rong Fan*, Jiangbing Zhou*, Cross-platform analysis reveals cellular and molecular landscape of glioblastoma invasion, Neuro-Oncology, 25 (3), 482-494 (2023)
Yanxiang Deng, Marek Bartosovic, Sai Ma, Di Zhang, Petra Kukanja, Yang Xiao, Graham Su, Yang Liu, Xiaoyu Qin, Gorazd B Rosoklija, Andrew J Dwork, J John Mann, Mina L Xu, Stephanie Halene, Joseph E Craft, Kam W Leong, Maura Boldrini, Gonçalo Castelo-Branco*, Rong Fan*, Spatial profiling of chromatin accessibility in mouse and human tissues, Nature, 609 (7926), 375-383 (2022).
Bai ZL, Woodhouse S, Kim D, Lundh S, Sun HX, Deng YX, Xiao Y, Barrett DM, Myers RM, Grupp SA, June CH, Melenhorst JJ*, Camara PG*, and Fan R*. Single-cell antigen-specific activation landscape of CAR T cell infusion product reveals determinates of CD19 positive relapse in patients with ALL, Science Advances, 8, eabj2820 (2022).
Enninful, A., Baysoy, A. & Fan, R*. Unmixing for ultra-high-plex fluorescence imaging. Nature Communication, 13, 3473 (2022).
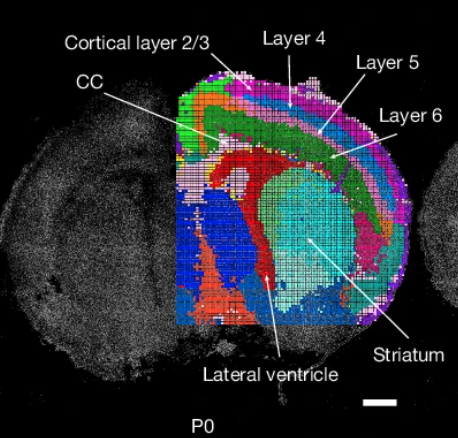
Deng Y, Bartosovic M, Kukanja P, Zhang D, Liu Y, Su G, Enninful A, Bai Z, Castelo-Branco G, Fan R*. Spatial- CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level. Science 375(6581):681-686 (2022).
Franjic D, Skarica M, Ma X, Arellano JI, Tebbenkamp ATN, Choi JM, Xu C, Li Q, Morozov YM, Andrijevic D, Vrselja Z, Spajic A, Santpere G, Li MF, Zhang S, Liu Y, Spurrier J, Zhang L, Gudelj I, Rapan L, Takahashi H, Huttner A, Fan R, Strittmatter SM, Sousa AMM, Rakic P, and Sestan N. Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells, Neuron, 110(3), 452-469.e14. (2022)
Jun Liu, Xiaoying Wang, Ann T. Chen, Xingchun Gao, Benjamin T. Himes, Hongyi Zhang, Zeming Chen, Jianhui Wang, Wendy C. Sheu, Gang Deng, Yang Xiao, Pan Zou, Shenqi Zhang, Fuyao Liu, Yong Zhu, Rong Fan, Toral R. Patel*, W. Mark Saltzman* & Jiangbing Zhou*, ZNF117 regulates glioblastoma stem cell differentiation towards oligodendroglial lineage. Nature Communications, 13, 2196 (2022).
Xiao Y, Wang Z, Zhao M, Deng Y, Yang M, Su G, Yang K, Qian C, Hu X, Liu Y, Geng L, Xiao Y, Zou Y, Tang X, Liu H, Xiao H, Fan R*. Single-Cell Transcriptomics Revealed Subtype-Specific Tumor Immune Microenvironments in Human Glioblastomas. Frontier in Immunology. 13: 914236 (2022)
Su G, Qin X, Enninful A, Bai Z, Deng Y, Liu Y, and Fan R. Spatial multi-omics sequencing for fixed tissue via DBiT- seq, STAR Protocols, 2, 100532 (2021).
Bai ZL, Lundh S, Kim D, Woodhouse S, Barrett DM, Myers RM, Grupp SA, Maus MC, June CH, Melenhorst JJ*, and Fan R*, Single cell multi-omics dissection of basal and antigen-specific activation states of CD19-targeted CAR T cells, Journal for ImmunoTherapy of Cancer (JITC), 9:e002328 (2021)
Yang Liu, Mingyu Yang, Yanxiang Deng, Graham Su, Archibald Enninful, Cindy C. Guo, Toma Tebaldi, Di Zhang, Dongjoo Kim, Zhiliang Bai, Eileen Norris, Alisia Pan, Jiatong Li, Yang Xiao, Stephanie Halene, and Rong Fan*, High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue, Cell, 183, 1665-1681 (2020).
Min Sun Shin, Dongjoo Kim, Kristina Yim, Hong-Jai Park, Sungyong You, Xuemei Dong, Fotios Koumpouras, Albert C.Shaw, Rong Fan, Smita Krishnaswamy, & Insoo Kang, IL-7 receptor alpha defines heterogeneity and signature of human effector memory CD8+ T cells in high dimensional analysis, Cellular Immunology, 355, 104155 (2020).
Gao Y, Vasic R, Song Y, Teng R, Liu C, Gbyli R, Biancon G, Nelakanti R, Lobben K, Kudo E, Liu W, Ardasheva A, Fu X, Wang X, Joshi P, Lee V, Dura B, Viero G, Iwasaki A, Fan R, Xiao A, Flavell RA, Li HB, Tebaldi T, and Halene S, m6A Modification Prevents Formation of Endogenous Double-Stranded RNAs and Deleterious Innate Immune Responses during Hematopoietic Development, Immunity, 52 (6), P1007-1021.E8 (2020).
Bai ZL, Deng YX, Kim D, Chen Z, Xiao Y, and Fan R*, Double-Sub-Poisson Single-Cell RNA-Sequencing via an Integrated DEP-Trapping and Nanowell Transfer Approach, ACS Nano, 14 (6), 7412–7424 (2020)
Finck A and Fan R*, Single-cell cytokine analysis of CAR-T cell activation. Cell Reprogramming for Immunotherapy pp 67-81 (2020).
Ding L, Su Y, Qiu X, Harper NW, Fassl A, Hinohara K, Huh SJ, Bloushtain-Qimron N, JovanovicB, Ekram M, Zi XY, Hines WC, Merino VF, Choudhury S, Ethington G, Panos L, Grant M, Rosson GD, Clark J, Herlihy W, Au A, Richardson AL, Argani P, Hwang ES, Fan R, Allred DG, Babski K, Kim EMH, McDonnell III CH, Wagner J, Rowberry R, Bobolis K, Kleer C, Blum JL, Sicinski P, Long HW, Sukumar S, Park SY, Garber JE, Bissell M, Yao J, Polyak K, Perturbed myoepithelial cell differentiation in BRCA mutation carriers and in ductal carcinoma in situ (DCIS) , Nature Communications, 10, 4182 (2019).
Xiao Y, Liu C, Chen Z, Blatchley MR, Zhou J, Xu M, Gerecht S, and Fan R*, Senescent cells with augmented cytokine production for microvascular bioengineering and tissue repairs, Advanced Biosystems, 3, 1900089 (2019). (Cover Highlight)
Janiszewska M, Tabassum DP, Castrano Z, Cristea S, Kingston NL, Ekram M, Harper NW, Kwak M, Qin Y, Laszewski T, Nakamura K, Luoma A, Marusyk A, Wucherpfennig KW, Fan R, Michor F, McAllister S, Polyak K, Subclonal cooperation drives metastasis through modulating local and systemic immune microenvironments, Nature Cell Biology, 21(7), 879 (2019).
Xhangolli I, Dura B, Lee GH, Kim DJ, Xiao Y, and Fan R*. Single-cell integrative analysis of CAR-T cell activation reveals a predominantly TH1/TH2 mixed response independent of differentiation, Genomics, Proteomics & Bioinformatics (GPB), 17(2), 129-139 (2019). (Cover Highlight)
Wang N, Zheng J, Chen Z, Liu Y, Dura B, Kwak M, Xavier-Ferrucio J, Lu Y-C, Zhang MM, Roden C, Cheng JJ, Krause D, Ding Y, Fan R* & Lu J*. Single-cell microRNA/mRNA co-profiling reveals non-(epigenetic) heterogeneity and regulatory programs. Nature Communications, 10: 95 (2019).
Chen Z, Lu Y, Zhang K, Xiao Y, Lu J, and Fan R*, Multiplexed, sequential secretion analysis of the same single cells reveals distinct effector response dynamics dependent on the initial basal state, Advanced Science, 6, 1801361 (2019). (Cover Highlight)
Chen Z, Chen JJ, and Fan R*. Single-cell protein secretion detection and profiling. Annual Review of Analytical Chemistry, 12:21.1-21.19, (2019).
Deng YX, Finck A, and Fan R*. Single-cell omics analyses enabled by microchip technologies. Annual Review of Biomedical Engineering, 21, 365-393 (2019).
Xiao Y, Kim DJ, Dura B, Zhang K, Yan RC, Li HM, Han E, Ip J, Zou P, Liu J, Chen AT, Vortmeyer AO, Zhou JB, and Fan R*, Ex vivo dynamics of human glioblastoma cells in a microvasculature-on-a-chip system correlates with tumor heterogeneity and subtypes, Advanced Science, 6, 1801531 (2019). (Cover Highlight)
Ren CG, Yuan QY, Braun M, Zhang X, Petri B, Zhang JS, Kim DJ, Guez-Haddad J, Xue WZ, Pan WJ, Fan R, Kubes P, Sun ZX, Opatowsky Y, Polleux F, Karatekin E, Tang WW, and Wu D, Leukocyte cytoskeleton polarization is initiated by plasma membrane curvature from cell attachment, Developmental Cell, 49, 1–14 (2019)
Adan Codina, Paul A Renauer, Guangchuan Wang, Ryan D Chow, Jonathan J Park, Hanghui Ye, Kerou Zhang, Matthew B Dong, Brandon Gassaway, Lupeng Ye, Youssef Errami, Li Shen, Alan Chang, Dhanpat Jain, Roy S Herbst, Marcus Bosenberg, Jesse Rinehart, Rong Fan, Sidi Chen, Convergent Identification and Interrogation of Tumor- Intrinsic Factors that Modulate Cancer Immunity In Vivo, Cell Systems, 8(2), 136-152.e7 (2019).
Wang NY, Lu Y, Chen Z, and Fan R*, Multiplexed PCR-Free Detection of MicroRNAs in Single Cancer Cells Using a DNA-Barcoded Microtrough Array Chip, Micromachines, 10(4), 215 (2019).
Dura B, Choi JY, Zhang K, Damsky W, Thakral D, Bosenberg M, Craft J, Fan R*, scFTD-seq: freeze-thaw lysis based, portable approach toward highly distributed single-cell 3’ mRNA profiling, Nucleic Acids Research, 47(3), e16 (2019).
Rossi J, Paczkowski P, Shen YW, Morse K, Flynn B, Kaiser A, Ng C, Gallatin K, Cain T, Fan R, Mackay S, Heath JR, Rosenberg SA, Kochenderfer JN, Zhou J, and Bot A, Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells associate with clinical outcomes in NHL, Blood, 132(8):804-814 (2018).
Xue Q, Bettini E, Paczkowski P, Ng C, Kaiser A, McConnell T, Kodrasi O, Quigley M, Heath JR, Fan R, Mackay S, Dudley M, Kassim S, and Zhou J, Single-cell multiplexed cytokine profiling of CD19 CAR-T cells revea ls a diverse landscape of polyfunctional antigen-specific response, Journal for ImmunoTherapy of Cancer, 5(1): 85 (2017).
Wang NY, Chen JJ, Fan R*, and Lu J*, Capture, Amplification, and Global Profiling of microRNAs from Low Quantities of Whole Cell Lysate, Analyst, 142, 3203-3211, (2017).
Phillip JM, Wu PH, Gilkes DM, Williams W, McGovern S, Daya J, Lee J. S.H., Chen JJ, Fan R, Walston J, Wirtz D, Biophysical and biomolecular determination of cellular age in humans, Nature Biomedical Engineering, 1, 0093 (2017).
Gil Del Alcazar, CR, Huh SJ, Ekram MB, Trinh A, Liu LL, Beca F, Zi XY, Kwak M, Bergholtz H, Su Y, Ding L, Russnes HG, Richardson AL, Babski K, EMH Kim, McDonnell C, Wagner J, Rowberry R, Freeman GJ, Dillon D, Sorlie T, Coussens LM, Garber JE, Fan R, Bobolis K, Allred DC, Jeong J, Park SY, Michor F and Polyak K, Immune Escape in Breast Cancer During In Situ to Invasive Carcinoma Transition, Cancer Discovery, 7(10):1098-1115, (2017)
Jayatilaka H, Tyle P, Chen JJ, Kwak M, Ju JA, Kim HJ, Lee J.S.H., WuPH, Gilkes DM, Fan R*, and Wirtz D*, Novel synergistic IL6/8 paracrine signaling pathway infers new strategy to inhibit tumor cell migration, Nature Communications, 8:15584, (2017).
Han L, Wu HJ, Zhu HY, Kim KY, Riester M, Marjani SL, Euskirchen G, Zi XY, Snyder M, Park IH, Weissman SM, Michor F*, Fan R* and Pan XH*, Bisulfite-independent analysis of CpG island methylation enables genome-scale stratification of single cells, Nucleic Acid Research, 45(10):e7, (2017).
Mao L, Shen L, Chen JH, Zhang XB, Kwak M, Wu Y, Fan R, Zhang L, Pei J, Yuan GY, Song CL, Ge JB & Ding WJ, A promising biodegradable magnesium alloy suitable for clinical vascular stent application, Scientific Reports, 7, 46343 (2017).
Liu J, Guo B, Chen Z, Wang N, Iacovino M, Cheng J, Roden C, Pan W, Chen S, Kyba M, Fan R, Guo S, and Lu J, MicroRNA-125b promotes MLL-AF9-driven murine acute myeloid leukemia involving a VEGFA-mediated non-cell- intrinsic mechanism, Blood, 129(11):1491-1502, (2017)
Chen JJ, Kwak M, and Fan R*. Single cell cytokine profiling to investigate cellular functional diversity in hematopoietic malignancies. Book Chapter: Chromic Myeloid Leukemia, Springer (2016).
Brower K and Fan R*, Cellular Immunophenotyping: Industrial Technologies and Emerging Tools. Book Chapter: Micro- and Nanosystems for Biotechnology. Wiley (2016).
Ching T, Peplowska K, Huang S, Zhu X , Shen Y, Molnar J, Yu H, Tiirikainen M, Fogelgren B, Fan R, and Garmire LX, Pan-cancer analyses reveal a panel of biologically and clinically relevant lincRNAs for tumour diagnosis, subtyping and prognosis, EBioMedicine, 7, 7-8 (2016).
Miller-Jensen K* and Fan R*, High-Throughput Secretomic Analysis of Single Cells to Assess Functional Cellular Heterogeneity, Experimental Approaches for the Investigation of Innate Immunity: The Human Innate Immunity Handbook. World Scientific (2016).
Kwak M, Han L and Fan R*., Interfacing inorganic nanowire arrays and living cells for cellular function analysis, Small, 42, 5600-5610 (2015).
Xue Q, Lu Y, Eisele MR, Sulistijo E, Fan R*, and Miller-Jensen K*, Analysis of single-cell secretion reveals a role for paracrine signaling in coordinating macrophage response to TLR4 stimulation, Science Signaling, 8, 381 (2015).
Gagliani N, Iseppon A, Vesely CA, Brockmann L, Palm NW, de Zoete MR, Licona-Limon P, Paiva P, Ching T, Weaver C, Zi X, Fan R, Garmire LX, Geginat J, Stockinger B, Esplugues E, Huber S, and Flavell RA*, Th17 cells transdifferentiate into regulatory cells during the resolution of inflammatory response, Nature, 523, 221–225 (2015).
Han L, Zhou J, Sun YB, Zhang Y, Han J, Fu JP, and Fan R*, Single-crystalline, Nanoporous Gallium Nitride Films with Fine Tuning of Pore Size for Stem Cell Engineering, ASME J. Nanotech. Eng. Med., 5(4), 041004 (2015).
Mao L, Shen L, Chen JH, Wu Y, Kwak M, Lu Y, Xue Q, Pei J, Zhang L, Yuan G*, Fan R*, Ge JB, and Ding WJ, Enhanced Bioactivity of Mg–Nd–Zn–Zr Alloy Achieved with Nanoscale MgF2 Surface for Vascular Stent Application, ACS Appl. Mater. Interfaces, 2015, 7 (9), pp 5320–5330
Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, Hricik T, Bhagwat N, Abdel-Wahab OI, Marubayashi S, Chen JJ, Romanet V, Fridman JS, Bromberg J, Murakami M, Radimerski T, Michor F, Fan R*, and Levine RL*. JAK-STAT Pathway Activation in Malignant and Non-Malignant Cells Contributes to MPN Pathogenesis and Therapeutic Response, Cancer Discovery, 5(3), 316–31 (2015).
Lu Y, Xue Q, Eisele MR, Sulistijo E, Brower K, Han L, Amir ED, Pe’er D, Miller-Jensen K *, and Fan R*, Highly multiplexed profiling of immune effector functions reveals deep functional heterogeneity in response to pathogenic ligands, Proc. Natl. Acad. Sci., U.S.A., 112(7), 607-615 (2015).
Han L, Zi XY, Garmire LX, Wu Y, Weissman SM*, Pan XH*, and Fan R*, Co-detection and sequencing of genes and transcripts from the same single cells facilitated by a microfluidics platform, Scientific Reports, 4, 6485 (2014).
Elitas M, Brower K, Lu Y, Chen JJ, and Fan R*, A microchip platform for interrogating tumor- macrophage paracrine signaling at the single-cell level, Lab on a Chip, 14, 3582 (2014).
Kwak M, Kim DJ, Han L, Lee MR, Wu Y, Lee SK, and Fan R*. Nanowire array chips for molecular typing of rare trafficking leukocytes with application to neurodegenerative pathology, Nanoscale, 6(12), 6537-50 (2014).
Guo S*, Zi XY, Schultz V, Cheng LJ, Zhong M, Koochaki S, Megyola CM, Pan XH, Heydari K, Weissman SM, Gallagher PG, Krause DS, Fan R, and Lu J, Non-stochastic reprogramming from a privileged somatic cell state, Cell, 156, 1-14 (2014).
Lee SK*, Kim DJ, Lee GH, and Fan R*, Specific rare cell capture using micro-patterned silicon nanowire platform, Biosensor and Bioelectronics, 54c, 181-188 (2014).
Kim DJ, Kim GS, Seol JK, Hyung JH, Park NW, Lee MR, Lee MK, Fan R, and Lee SK*, Filopodial Morphology Correlates to the Capture Efficiency of Primary T-Cells on Nanohole Arrays, Journal of Biomedical Nanotechnology, 10, 1030-1040 (2014).
Mao L, Shen L, Niu J, Zhang J, Fan R*, and Yuan GY*. Nanophasic biodegradation enhances the durability and biocompatibility of magnesium alloys for the next-generation vascular stents. Nanoscale, 5, 9517-9522 (2013).
Wei W, Shin YS, Ma C, Wang J, Elitas M, Fan R, and Heath JR*, Microchip platforms for multiplex single-cell functional proteomics with applications to immunology and cancer research, Genome Medicine, 5, 75 (2013)
Jonas S, Zhou E, Deniz E, Huang B, Chandrasekera K, Bhattacharya D, Wu Y, Fan R, Deserno TM, Khokha MK, and Choma MA*, A novel approach to quantifying ciliary physiology: microfluidic mixing driven by a ciliated biological surface, Lab on a Chip, 13, 4160-4163 (2013)
Shomorony A and Fan R*, Immuno-DNA-directed Assembly of Multicellular Systems, Chemistry Letters, 42 (5), 512-514 (2013).
Lu Y, Chen JJ, Mu L, Xue Q, Wu Y, Wu PH, Li J, Vortmeyer AO, Miller-Jensen K, Wirtz D, and Fan R*, High- throughput Secretomic Analysis of Single Cells to Assess Functional Cellular Heterogeneity, Analytical Chemistry, 85 (4), 2548–2556, (2013).
Chen W, Huang NT, Oh B, Lam RHW, Fan R, Cornell TT, Shanley TP, Kurabayashi K, and Fu JP, Surface- micromachined microfiltration membranes for efficient isolation and functional immunophenotyping of subpopulations of immune cells, Advanced Healthcare Materials, 2: 921, 2013.
Ma C*, Fan R* and Elitas M, Single cell functional proteomics for assessing immune response in cancer therapy: technology, methods, and applications, (review article) Frontiers in Oncology, 3:133 (2013)
Kwak M, Mu L, Lu Y, Chen JJ, Brower K, and Fan R*, Single-cell Protein Secretomic Signatures as Potential Correlates to Tumor Cell Lineage Evolution and Cell-cell Interaction. Frontiers in Oncology, 3:10 (2013).
Wu Y, Garmire LX and Fan R*, Intercellular Signaling Network Reveals a Mechanistic Transition in Tumor Microenvironment, Integrative Biology, 4, 1478-1486, (2012).
Zhou J , Wu Y, Lee SK and Fan R*, High-content single cell analysis on chip using a laser microarray scanner, Lab on a Chip, 12, 5025-5033 (2012).
Lee SK, Kim, GS, Wu Y, Lu Y, Han L, Kim DJ, Hyung JH, Seol JK, Sander C, Gonzalez A, Li J & Fan R*, Nanowire Substrate-based Laser Scanning Cytometry for Quantitation of Circulating Tumor Cells, Nano Letters, 12(6), 2697- 2704 (2012)
Chen WQ, Villa-Diaz LG, Weng SN, Kim JK, Lam R, Han L, Fan R, Krebsbach PH, and Fu JP*, Nanotopography influences adhesion, spreading, and self-renewal of human embryonic stem cells, ACS Nano, 6, 4094-4103 (2012).
Kim DJ, Seol JK, Wu Y, Ji S, Kim GS, Hyung JH, Lee SY, Im H, Fan R* and Lee SK*, Quartz nanopillar hemocytometer for high-yield separation and counting of CD4+ T lymphocytes, Nanoscale, 4, 2500-2507 (2012). (cover highlight)
Shi QH, Qin LD, Fan R, Wei W, Guo DL, Shin YS, Hood L, Mischel P, and Heath JR, Single cell proteomic chip for profiling intracellular signaling pathways in single tumor cells, Proc. Nat. Acad. Sci., U.S.A. 109(2), 419-424 (2012).
Wu Y, Lu Y, Chen WQ, Fu JP, and Fan R*. In silico experimentation of glioma microenvironment development and anti-tumor therapy. PLoS Computational Biology, 8(2), 1002355 (2012).
Guan WH, Fan R, Reed MA, Field effect reconfigurable nanofluidic diodes, Nature Communication, 2, 506 (2011).
Ma C#, Fan R #, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A and Heath JR. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells, Nature Medicine, 17, 738-743 (2011). (# equal contribution)
Shin YS, Remacle F, Fan R, et al. Protein signaling networks from single cell fluctuations and information theory profiling, Biophysical Journal, 100, 2378-2386 (2011).
Fan R#, Vermesh, O.#, Srivastava, A., Yen, B.K.H., Qin, L.D., Ahmad, H., Kwong, G.A., Liu, C.C., Gould, J., Hood, L. & Heath, J.R., Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nature Biotechnology 26, 1373-1378 (2008). (# co-first author). This article is highlighted by the cover illustration.
Vermesh, U., et al. Fast Nonlinear Ion Transport via Field-induced Hydrodynamic Slip in Sub-20-nm Hydrophilic Nanofluidic Transistors. Nano Letters 9, 1315-1319 (2009).
Fan R, Huh S, Yan R, Arnold J and Yang PD. Gated proton transport in aligned mesoporous silica films. Nature Materials 7, 303-307 (2008).
Sirbuly, D.J., Tao, A., Law, M., Fan, R. & Yang, P.D. Multifunctional nanowire evanescent wave optical sensors. Advanced Materials 19, 61-+ (2007).
Huang, J.X., Fan, R., Connor, S. & Yang, P.D. One-step patterning of aligned nanowire arrays by programmed dip coating. Angewandte Chemie-International Edition 46, 2414-2417 (2007).
Zhang, Y., et al. Characterization of heat transfer along a silicon nanowire using thermoreflectance technique. IEEE Transactions on Nanotechnology 5, 67-74 (2006).
Gui, Z., et al. Reply to "Comment on precursor morphology controlled formation of rutile VO2 nanorods and their self-assembled structures". Chemistry of Materials 18, 5630-5630 (2006).
Goldberger, J., Hochbaum, A.I., Fan, R. & Yang, P.D. Silicon vertically integrated nanowire field effect transistors. Nano Letters 6, 973-977 (2006).
Goldberger, J., Fan, R. & Yang, P.D. Inorganic nanotubes: A novel platform for nanofluidics. Accounts of Chemical Research 39, 239-248 (2006).
Karnik, R.*, Fan, R.*, Yue, M., Li, D.Y., Yang, P.D. & Majumda, A. Electrostatic control of ions and molecules in nanofluidic transistors. Nano Letters 5, 943-948 (2005). (*co-first)
Karnik, R., Castelino, K., Fan, R., Yang, P. & Majumdar, A. Effects of biological reactions and modifications on conductance of nanofluidic channels. Nano Letters 5, 1638-1642 (2005).
Hochbaum, A.I., Fan, R., He, R.R. & Yang, P.D. Controlled growth of Si nanowire arrays for device integration. Nano Letters 5, 457-460 (2005).
He, R.R., et al. Si nanowire bridges in microtrenches: Integration of growth into device fabrication. Advanced Materials 17, 2098-+ (2005).
Fan, R., Yue, M., Karnik, R., Majumdar, A. & Yang, P.D. Inorganic nanotube nanofluidic transistors for single molecule detection. ACS Annul Meeting Abstract, 323-ANYL (2005).
Fan, R., Yue, M., Karnik, R., Majumdar, A. & Yang, P.D. Polarity switching and transient responses in single nanotube nanofluidic transistors. Physical Review Letters 95(2005).
Fan, R., Karnik R., Yue, M., Majumda, A. & Yang P.D. DNA translocation in inorganic nanotubes. Nano Letters 5, 1633-1637 (2005).
Wu, B.M., et al. Two-bandgap magneto-thermal conductivity of polycrystalline MgB2. Superconductor Science & Technology 17, 1458-1463 (2004).
Gui, Z., Fan, R., Chen, X.H., Hu, Y. & Wang, Z.Z. A new colloidal precursor cooperative conversion route to nanocrystalline quaternary copper sulfide. Materials Research Bulletin 39, 237-241 (2004).
Yang, D.S., et al. Thermal conductivity of two-energy-gap superconductor MgB2. Acta Physica Sinica 52, 683- 686 (2003).
Sun, Z., Chen, X.H., Fan, R., Luo, X.G. & Li, L. Structure and magnetic properties of perovskite Sr2CuNbO6-75. Journal of Physics and Chemistry of Solids 64, 59-62 (2003).
Li, D.Y., Wu, Y., Fan, R., Yang, P.D. & Majumdar, A. Thermal conductivity of Si/SiGe superlattice nanowires. Applied Physics Letters 83, 3186-3188 (2003).
Gui, Z., et al. Synthesis and characterization of reduced transition metal oxides and nanophase metals with hydrazine in aqueous solution. Materials Research Bulletin 38, 169-176 (2003).
Fan, R., Wu, Y.Y., Li, D.Y., Majumda, A. & Yang, P.D. Fabrication of silica nanotube arrays from vertical silicon nanowire templates. Journal of the American Chemical Society 125, 5254-5255 (2003).
Zhang, H.T., Gui, Z., Fan, R. & Chen, X.H. Hydrothermal synthesis and characterization of nanorods "LixV2- delta O4-delta center dot H2O". Inorganic Chemistry Communications 5, 399-402 (2002).
Wu, Y.Y., Fan, R. & Yang, P.D. Block-by-block growth of single-crystalline Si/SiGe superlattice nanowires. Nano Letters 2, 83-86 (2002). Notable coverage:
Wang, C.H., et al. Transport properties of new superconductor MgCNi3. Acta Physica Sinica 51, 1816-1820 (2002).
Li, S.Y., et al. Thermopower and thermal conductivity of superconducting perovskite MgCNi3. Physical Review B 65(2002).
He, R.R., Law, M., Fan, R., Kim, F. & Yang, P.D. Functional bimorph composite nanotapes. Nano Letters 2, 1109-1112 (2002).
Gui, Z., et al. Precursor morphology controlled formation of rutile VO2 nanorods and their self-assembled structure. Chemistry of Materials 14, 5053-5056 (2002).
Li, Y.P., Fang, Y.E., Fan, R. & Xing, J.Y. Study of plasma-polymerization deposition of C2H2/CO2/H-2 onto ethylene-co-propylene rubber membranes. Radiation Physics and Chemistry 60, 637-642 (2001).
Li, S.Y., et al. Alkali metal substitution effects in Mg(1-x)A(x)B(2) (A = Li and Na). Physica C 363, 219-223 (2001).
Li, S.Y., Liu, L., Fan, R. & Chen, X.H. Synthesis and superconductivity study for (Hg,M)-1222 phase (M = W, Mo, V, Cr, Ti). Physica C 356, 192-196 (2001).
Li, S.Y., Fan, R., et al. Normal state resistivity, upper critical field, and Hall effect in superconducting perovskite MgCNi3. Physical Review B 64(2001).
Li, H.L., et al. Study on the resistivity and Hall effect of MgB2 and Mg0.93Li0.07B2. Acta Physica Sinica 50, 2044-2048 (2001).
Gui, Z., Fan, R., Chen, X.H. & Wu, Y.C. A new metastable phase of needle-like nanocrystalline VO2 center dot H2O and phase transformation. Journal of Solid State Chemistry 157, 250-254 (2001).
Gui, Z., Fan, R., Chen, X.H. & Wu, Y.C. A simple direct preparation of nanocrystalline gamma-Mn2O3 at ambient temperature. Inorganic Chemistry Communications 4, 294-296 (2001).
Gui, Z., Fan, R., Chen, X.H., Hu, Y. & Wu, Y.C. Hydrothermal synthesis and magnetic property of NaV2O5 nanorods. Transactions of Nonferrous Metals Society of China 11, 324-327 (2001).
Fan, R., Chen, X.H., Gui, Z., Liu, L. & Chen, Z.Y. A new simple hydrothermal preparation of nanocrystalline magnetite Fe3O4. Materials Research Bulletin 36, 497-502 (2001).
Fan, R., Chen, X.H., Gui, Z., Li, S.Y. & Chen, Z.Y. Sim-deintercalation synthesis and electronic conduction of hexagonal potassium tungsten bronze. Materials Letters 49, 214-218 (2001).
Chen, X.H. & Fan, R. Low-temperature hydrothermal synthesis of transition metal dichalcogenides. Chemistry of Materials 13, 802-805 (2001).
Fan, R., et al. Chemical synthesis and electronic conduction properties of sodium and potassium tungsten bronzes. Journal of Physics and Chemistry of Solids 61, 2029-2033 (2000).
Fan, R., Chen, X.H. & Chen, Z.Y. A novel route to obtain molybdenum dichalcogenides by hydrothermal reaction. Chemistry Letters, 920-921 (2000).